Sanger sequencing vs. NGS sequencing
I've always wondered about the differences. Sin Lee explained to me on the phone today. I'll explain why this is important. Wondering what variant you have? We'll explain how to find out.
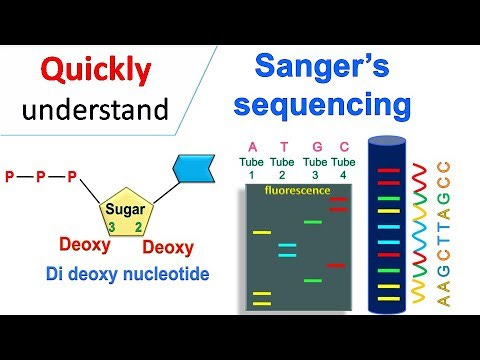
Sanger sequencing
Sanger sequencing allows a lab to find the DOMINANT strain of SARS-CoV-2 in a sample. Two labs with the exact same sample should always find EXACTLY the same genetic sequence. This is important because a person might have 100 variants of the genome. This test finds the genomic sequence that is the most popular real, existing sequence.
NGS gene sequencing
NGS sequencing will the find the CONSENSUS sequence of the SARS-CoV-2 variants in a given sample. So at each position in the gene, it will find the nucleotide that is the most popular in the sample at that position.
This is important: the consensus sequence may not even physically exist in nature as a real physical single strand, but it is a way to characterize the mix of variants in a person.
Two labs with identical samples will find APPROXIMATELY the same consensus sequence but not identical.
This is because there is noise in the reader and the sample and if the nucleotides are evenly matched (e.g., even numbers of A and T for example), it could be random what the result is for that position. It’s like an election with four candidates and two of them have a tie for first place.
Why this matters
People who claim the virus doesn’t exist will say the NGS sequencing produces random results. That’s disingenuous. It doesn’t do that at all.
The consensus sequences are unique to a person because most people will have dozens of variants. The NGS sequences are like a person’s fingerprints since no two people will have exactly the same mix of variants. And your variant mix will vary over time. So your NGS sequence today won’t necessarily be the same as tomorrow.
Therefore, if I infect my wife with my virus, her NGS sequence can be quite different than mine. This is because her body will make unique mutations of the virus internally and her body will amplify different variants than mine (e.g., she was vaccinated and I wasn’t or she had different colds than I had).
Therefore, our NGS sequences WILL be different even if we were infected with exactly the SAME variant(s) of the virus.
Our Sanger sequences may or may not be the same since again, she is getting lots of variants and may amplify them differently. Sanger picks the one she has the most copies of.
What is true is that two samples from the same patient taken at exactly the same time will have IDENTICAL Sanger sequences and nearly identical NGS sequences.
The right way to confirm a virus
In the old days, we’d specify things correctly. For example, from Nucleic Acid Amplification Assay for the Detection of Enterovirus RNA - Class II Special Controls Guidance for Industry and FDA Staff, we have:
Detection of an EV genome in CSF by two different well-characterized and validated nucleic acid amplification tests (NAAT). The NAAT primers pairs should generate amplicons from different genomic regions. One of the NAAT assays should provide sequence information. Bi-directional sequencing should be performed on both strands of the amplicon and the generated sequence should be of an acceptable quality (quality score of 40 or higher as measured by PHRED or similar software packages) and should match the reference or consensus sequence [Ref. 10, 17].
Those days are gone. Today, the recommendations for COVID are really watered down to reduce costs, resulting in lots of false positives and negatives.
Want to find out which variant you are infected with?
I highly recommend Sin Lee’s lab
The test costs $150 and you get lots of information because if you get a “hit” they will do further sequences at no additional cost.
Summary
For finding the dominant strain of virus in a person, Sanger sequencing is the gold standard. A test for SARS-CoV-2 costs around $100.
If you are trying to characterize the range of SARS-CoV-2 viral genome present in a person, you’d want to use the consensus genome obtained from NGS.
The sequences will be different for a given sample.
I hope this helps clear things up.


If I infect my wife with my virus, my MAN sequence is to order flowers, take out the trash and cook dinner.
But seriously, this does clear things up, thanks!
WTF? You've skipped step 1; what is the "sample" and what was the process for obtaining it? We need to see the steps.
Sequencing soup of a cell culture that includes animal cell lines, antibiotics and more tells you nothing